Powered by Chemaxon™, chemical names have been extracted from the entire Fampat Database
How to search?
There are two ways to enter a search: typing a text or drawing a molecule.
You can also limit your molecule searches to specific fields:
• Title
• Abstract
• Claims
• Description
• Images/.mol files
Search by text
Supported names include
•Common names (e.g. Toluene, Aspirin)
•Common drug names (e.g. Paracetamol, Doliprane)
•Acronyms (e.g. ATP for "Adenosine Triphosphate")
•CAS numbers
•IUPAC names, CAS names and generally systematic names
•SMILES codes
Current limitations
•Molecules are extracted from CA, FR, DE, IL, EP, WO, US, KR, JP, GB, AU, CN, IN publications
•Molecules inside additional.mol files are only extracted from 2007 US publications.
•Molecules are extracted from images inside US, WO, EP et JP publications from 2007.
•Names containing isotopes are not supported yet.
Note: the search is case sensitive (CO is different from Co), acronyms have to be searched in upper cases.
Exact structure search
Choosing the exact search option will search for the exact molecule without any variation on the structure.
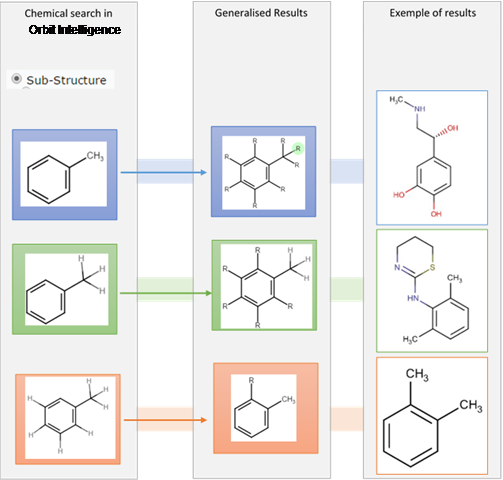
Sub-structure search
Choosing the sub-structure search option will search for the molecule allowing for the replacement of any hydrogens with any atoms.
Drawing out manually a hydrogen will render this hydrogen not replaceable even in sub-structure search.
Note: this option must be chosen for any R group even precisely defined
You can read the online Help Links for drawing (Marvin JS). For a start: Getting Started with Marvin JS
If you want to go further, you can find here a documentation about the sketcher options, and detailed information about drawing R-groups in Marvin JS.

If you are interested in the query features, which can be drawn in Marvin JS, a summary page is at your disposal.
Differences between a structure with hydrogens drawn out and hydrogens implied in the sub structure search:
In the sub-structure search if you do not actively draw out a hydrogen on a carbon it is considered by the system as a variation point in your sub-structure search. If you purposely draw the hydrogen on the carbon you lock that position and force the system to search the structure with a hydrogen on that carbon.
Principle is explained
in the graph below.
Q&A
If I choose the text search over the draw option and type aspirin would the system search for acetylsalicylic acid?
ü Yes. Chemaxon™ searches for chemical names and merge all names corresponding to the same molecule. That includes commercial names, common names, IUPAC names…
If I’m using the Markush drawing search module ?
ü It is useful to know that we can use Markush drawing as a drawing search mode. However, Markush structures are not indexed, here are some example of the use of Markush search
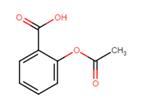
Example 1: let’s take the example of a patent discussing ‘aspirin’. Indexation of the Chemistry Module will transform the word aspirin into the structure below.
It is possible for a user to draw a Markush structure (see below) to search a family of structures that include aspirin and find all the resulting documents (including aspirin).
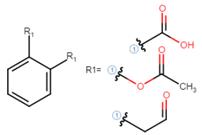
Markush drawing above can be drawn to search 6 molecules at once (removing symetric compounds), one of which being aspirin.
Example 2: limitations with the following text “The quinoline 36 was modified with an acid moiety and a halogen group to obtain various structures such as 7-iodo-6-quinoline-acetic acid.”
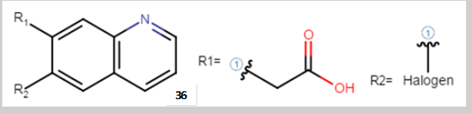
In the above Markush structure, we can see two R groups: R1 and R2.
R is used as a variation point. The variation is then explained, R1 is an acid moiety while R2 can be any atom of the halogen group.
Therefore structure 36 is actually representing 5 different molecules which can be drawn out as follow:
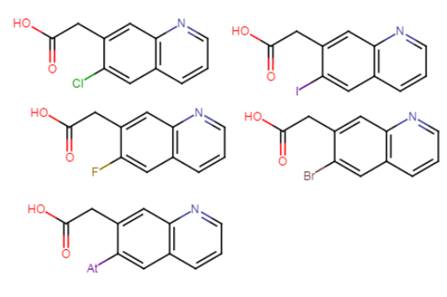
We have replaced the R1 with the acid moiety and the R2 with all possible halogen atoms.
How do we index it in Orbit?
ü Additional indexed molecules have been extracted from images for US, WO, EP and JP after 2007 and from .mol files for US, also starting from 2007. .
ü The structure “7-iodo-6-quinoline-acetic acid” will be detected even if published for the first time. The naming follows chemical naming rules and thus can be read by Orbit Intelligence.
ü The text: ‘quinoline 36 was modified with an acid moiety and a halogen group will be detected as quinoline, acid, halogen:
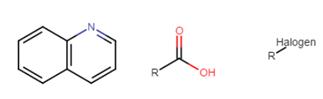
Therefore, searching
for the molecule below will not retrieve the line “quinoline 36 was modified
with an acid moiety and a halogen group” but will retrieve
“7-iodo-6-quinoline-acetic acid”. 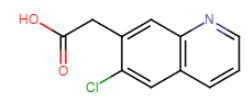
Note: searching for the molecule below in the sub-structure search mode (i.e. allowing variation on all carbon) will retrieve quinoline 36.
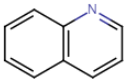 The patent will then be
retrieved.
The patent will then be
retrieved.
What format can I use to search in Orbit Intelligence?
ü CAS numbers can be used to search patents as they will be converted to chemical names. However, CAS numbers are not stored and thus cannot be found in patent data.
ü SMILES can be used to search molecules, e.g. C1=CC2=C(C=CC(=C2)C(=O)O)N=C1
ü IUPAC names can be detected.
ü Pharmaceuticals trade names.